 This study investigates the maternal lineage diversity, genetic relationships, and demographic history of six Tanzanian indigenous chicken ecotypes, including Sukuma, Kuchi, Unguja, Pemba, Morogoro Medium, and Ching’wekwe chicken ecotypes using mitochondrial DNA (mtDNA) D-loop analysis. Indigenous chickens play a vital role in rural livelihoods, food security, and cultural practices in Tanzania, yet they face increasing threats from genetic erosion caused by uncontrolled crossbreeding and the introduction of exotic breeds.
This study investigates the maternal lineage diversity, genetic relationships, and demographic history of six Tanzanian indigenous chicken ecotypes, including Sukuma, Kuchi, Unguja, Pemba, Morogoro Medium, and Ching’wekwe chicken ecotypes using mitochondrial DNA (mtDNA) D-loop analysis. Indigenous chickens play a vital role in rural livelihoods, food security, and cultural practices in Tanzania, yet they face increasing threats from genetic erosion caused by uncontrolled crossbreeding and the introduction of exotic breeds.
A total of 100 mtDNA D-loop sequences (310 bp) were analyzed, including newly generated sequences from Sukuma chickens and comparative sequences from other ecotypes retrieved from GenBank. Molecular diversity indices, phylogenetic analyses, analysis of molecular variance (AMOVA), and neutrality tests were applied to assess population structure and evolutionary dynamics. The results revealed strong and significant genetic differentiation among ecotypes, with 35.45% of variation occurring between populations and 64.55% within populations. Sukuma chickens exhibited the highest haplotype and nucleotide diversity, indicating multiple maternal origins and a long evolutionary history, while Kuchi chickens showed very low diversity, consistent with founder effects and intensive selection.
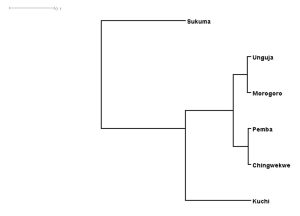 Phylogenetic analysis demonstrated clear clustering of inland ecotypes (Sukuma, Kuchi, Morogoro Medium), whereas coastal and island ecotypes (Unguja, Pemba, Ching’wekwe) showed closer genetic relationships, reflecting historical and contemporary gene flow linked to trade and human movement. Neutrality tests suggested demographic stability for most ecotypes, with limited evidence of recent population expansion.
Phylogenetic analysis demonstrated clear clustering of inland ecotypes (Sukuma, Kuchi, Morogoro Medium), whereas coastal and island ecotypes (Unguja, Pemba, Ching’wekwe) showed closer genetic relationships, reflecting historical and contemporary gene flow linked to trade and human movement. Neutrality tests suggested demographic stability for most ecotypes, with limited evidence of recent population expansion.
 Overall, the findings highlight Sukuma chickens as a key genetic resource for conservation-oriented breeding programs, while emphasizing the urgent need for genetic management strategies for low-diversity ecotypes such as Kuchi. The study provides a strong scientific foundation for designing sustainable conservation and breeding strategies to safeguard Tanzania’s indigenous chicken genetic heritage.
Overall, the findings highlight Sukuma chickens as a key genetic resource for conservation-oriented breeding programs, while emphasizing the urgent need for genetic management strategies for low-diversity ecotypes such as Kuchi. The study provides a strong scientific foundation for designing sustainable conservation and breeding strategies to safeguard Tanzania’s indigenous chicken genetic heritage.
For more information read:
Mhando, Z.M., Mbaga, S.H., Nguluma, A.S., Mwega, E.D. and Lyimo, C.M. (2026). Maternal Lineage Diversity and Genetic Relationships of Sukuma Chicken Ecotype with Other Tanzanian Indigenous Chickens Based on Mitochondrial DNA D-loop Region. Front. Anim. Sci., Sec. Animal Breeding and Genetics. 6:1732717, doi: 10.3389/fanim.2025.1732717

The Department of Animal, Aquaculture, and Range Sciences
The College of Agriculture, Sokoine University of Agriculture
Share this page

